wwPDB 2013 News
Contents
12/13/2013
Online PDBx/mmCIF Resources
The wwPDB has established PDBx/mmCIF as the new standard format for data exchange and archiving in structural biology
(more).
To help facilitate the transition from PDB to PDBx/mmCIF format, the wwPDB has created a new website
hosting PDBx/mmCIF Resources at mmcif.wwpdb.org.
11/27/2013
New wwPDB service allows crystallographers to validate their own structures
The Worldwide Protein Data Bank (wwPDB) partners are pleased to announce that X-ray structure validation reports can now be generated on demand by macromolecular crystallographers by using the new stand-alone wwPDB validation server.
The reports can be used to assess the quality of early, intermediate and near-final models to identify any potential problems that need addressing prior to structure analysis, publication and deposition.
"The stand-alone validation server will run exactly the same validation tests that have recently been introduced for the annotation of new depositions," says Randy Read of Cambridge University. Read chairs the wwPDB X-ray Validation Task Force (VTF) that has been active since 2008 and has produced detailed recommendations to the wwPDB about how macromolecular crystal structures should be validated [1]. Many of these recommendations have already been implemented by wwPDB in a software pipeline [2] that has been in use since August 2013 to validate new depositions. "With the stand-alone server, crystallographers won't have any last-minute surprises when they deposit their structures just before submitting the paper," Read adds.
Gerard Kleywegt was a member of the X-ray VTF until he moved to the EMBL-EBI in Cambridge in 2009, where he heads up the Protein Data Bank in Europe (PDBe), one of the wwPDB partner organisations. One of the tasks in his new job is to oversee the implementation of the validation pipelines for X-ray, NMR and EM structural data and models. Kleywegt explains: "The X-ray VTF was eager for wwPDB to offer a publicly accessible stand-alone version of the validation pipeline. We think this will help crystallographers by enabling them to identify any problems with their models or data before they even write their papers or deposit. Ultimately, the goal is to improve the overall quality of structures described in the literature and deposited to the relevant archives, PDB, EMDB and BMRB." Implementation of additional validation pipelines for structures based on NMR [3] and EM [4] data is in progress and these will be made publicly accessible in the future.
The stand-alone validation server was developed in the context of a larger initiative, the new wwPDB Deposition and Annotation System, which was created to unify the annotation tools and practices used across all wwPDB deposition centres. The new deposition system, currently in testing, will support all experimental methods currently archived by the wwPDB. The new system incorporates tools for validation of deposited structures as well as all of the chemical, sequence, structure and administrative annotation tasks. Helen Berman, director of the wwPDB deposition centre at the RCSB PDB, says: "By providing access to the same tools used by the wwPDB to validate crystallographic data, the validation server will become an important tool for our depositors."
The X-ray validation pipeline produces reports that include results from tried and tested software, including MolProbity, Xtriage, Mogul, EDS and various CCP4 programs. The reports include the results of geometric checks, structure-factor assessment, and ligand validation. They summarise the quality of the structure and highlight specific concerns by considering the coordinates of the model, the diffraction data and the fit between the two. Easily interpretable summary information that compares the quality of a structure with that of other structures in the archive is also provided.
We welcome feedback on the new validation reports and the stand-alone server. If you would like to send us your comments or questions, then please contact validation@mail.wwpdb.org
[1] Read R. J., Adams P. D., Arendall III W. D., Brünger A. T., Emsley P., Joosten R. P., Kleywegt G. J., Krissinel E. B., Lütteke T., Otwinowski Z., Perrakis A., Richardson J. S., Sheffler W. H., Smith J. L., Tickle I. J., Vriend G., and Zwart P. H. A new generation of crystallographic validation tools for the Protein Data Bank. Structure, 19, 1395-1412, 2011. DOI: 10.1016/j.str.2011.08.006
[2] Gore S., Velankar S. and Kleywegt G. J. Implementing an X-ray validation pipeline for the Protein Data Bank. Acta Cryst., D68, 478-483, 2012. DOI: 10.1107/S0907444911050359
[3] Montelione G. T., Nilges M., Bax A., Güntert P., Herrmann T., Richardson J. S., Schwieters C. D., Vranken W. F., Vuister G. W., Wishart D. S., Berman H. M., Kleywegt G. J. and Markley J .L. Recommendations of the wwPDB NMR Validation Task Force. Structure, 21, 1563-1570. DOI: 10.1016/j.str.2013.07.021
[4] Henderson R., Sali A., Baker M. L., Carragher B., Devkota B., Downing K. H., Egelman E., Feng Z., Frank J., Grigorieff N., Jiang W., Ludtke S. J., Medalia O., Penczek P. A., Rosenthal P. B., Rossmann M. G., Schmid M. F., Schröder G. F., Steven A. C., Stokes D. L., Westbrook J. D., Wriggers W., Yang H., Young J., Berman H. M., Chiu W., Kleywegt G. J., Lawson C. L. Outcome of the First Electron Microscopy Validation Task Force Meeting. Structure, 20, 205-214, 2012. DOI: 10.1016/j.str.2011.12.014

Randy Read at the inaugural wwPDB X-ray VTF meeting at the EMBL-EBI in 2008.
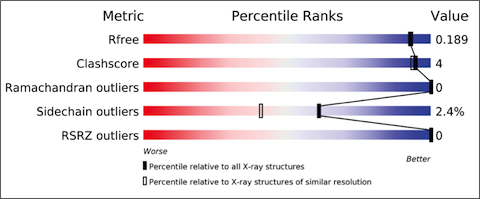
The validation reports provide at-a-glance summary information that compares the quality of a model with that of other models in the archive.
10/01/2013
Workshop on the PDBx/mmCIF Data Exchange Format For Structural Biology
Center For Integrative Proteomics Research
Rutgers, The State University of New Jersey
Tuesday October 22, 2013
http://www.proteinmodelportal.org/workshop-2013/
The wwPDB has established PDBx/mmCIF as the new standard format for data exchange and archiving in structural biology.
To help facilitate the transition from PDB to PDBx/mmCIF format the wwPDB is organizing a programmer's workshop
describing the content and organization of PDBx/mmCIF data, and the available software tools and libraries supporting PDBx/mmCIF (C/C++, Java, and Python).
The workshop will be held on Tuesday, October 22, 2013 at Rutgers, The State University of New Jersey
(in conjunction with a PSI Workshop on Theoretical Model Archiving and Validation to be held Monday, October 21, 2013):
Starting at 9am, the morning session will include presentations from:
Paul Adams (Phenix), David Case (AMBER), Tom Goddard (UCSF Chimera),
Robert Hanson (JMol), Eugene Krissinel (CCP4/mmdb), Andreas Prilic (BioJava), John Westbrook (RCSB PDB).
The afternoon session will offer an opportunity to work hands-on with
the presenters and developers in the areas of structural biology and
molecular modeling to learn application details and to discuss
successful approaches and experiences in adapting software to support PDBx/mmCIF.
In addition, the workshop will include a discussion of PDBx/mmCIF extensions required to represent molecular modeling specific information.
There is no registration fee, but online registration is required
(registration Deadline is October 4). For detailed agenda and online registration,
visit http://www.proteinmodelportal.org/workshop-2013/.
09/03/2013
Paper Published
How Community Has Shaped the Protein Data Bank
Helen M. Berman, Gerard J. Kleywegt, Haruki Nakamura, John L. Markley
Structure (2013) 21: 1485-1491 doi: 10.1016/j.str.2013.07.010
09/03/2013
wwPDB NMR Validation Task Force Recommendations Published
We are pleased to announce that the recommendations of the wwPDB
NMR Validation Task Force have been published in the journal Structure
(doi: 10.1016/j.str.2013.07.021).
As the number of structures in the PDB determined using NMR continues to grow, the provision of robust validation tools is becoming increasingly important. Assessing the quality of NMR structures and underlying experimental data is a critical area of NMR methods development, and also an essential component
of the process of making NMR structures accessible and useful to the wider scientific community.
The wwPDB partners are actively driving the development of new standards and software for the validation of structures determined by NMR spectroscopy. A wwPDB NMR Validation Task Force (VTF) comprised of experts in the field has been convened to define standard validation criteria, representing community consensus. The VTF has provided its initial recommendations, which will be implemented in a wwPDB validation pipeline and applied to all depositions of NMR structures.
This paper summarizes the recommendations of the NMR VTF, and lays the groundwork for future work in developing standards and metrics for biomolecular NMR structure and data quality assessment.
08/02/2013
New wwPDB X-ray structure validation reports support depositors, journal editors and referees.
The wwPDB partners are pleased to announce that new, considerably more informative X-ray structure validation reports are now being provided to depositors as part of the structure annotation process. The reports can be used by depositors to assess the quality of their structures, and by journal editors and referees as a useful tool in the manuscript-review process.
The new reports implement recommendations of a large group of community experts on validation [1,2]. They were originally scheduled to be introduced as part of the new wwPDB Deposition & Annotation system, which will come online in 2014. However, initial feedback on the reports has been very positive and therefore the reports are provided for all new X-ray crystal structures deposited PDBe, PDBj, and RCSB PDB as of August 1, 2013.
"Validation 'at the gate' is crucial to improving the quality and consistency of the structural archive," said Gerard Kleywegt, Head of PDBe. "The new wwPDB validation reports for X-ray crystal structures draw on a wealth of community experience in the validation of models, experimental data and the fit of the model to these data. The new style reports are a huge improvement compared to the reports previously provided by the wwPDB partners. Initial feedback from depositors, annotation staff and users alike has been very positive and this has in fact motivated us to start delivering the reports much earlier than we had planned. We strongly encourage journal editors and referees to request these reports from depositors as part of the manuscript submission and review process."
Provision of wwPDB validation reports is already required by the International Union of Crystallography (IUCr) journals as part of their submission process, and the wwPDB partners hope that the launch of the new improved reports will encourage other journals to follow suit.
The new X-ray validation reports have been prepared according to the recommendations of the wwPDB X-ray Validation Task Force [1,2], first convened in April 2008. The validation pipeline uses tried and tested software, including MolProbity, Xtriage, Mogul, EDS and various CCP4 programs. The reports include the results of geometry checks, structure factor validation, and ligand validation. They summarise the quality of the structure and highlight specific concerns by considering the coordinates of the model, the diffraction data and the fit between the two. Easily interpretable summary information that compares the quality of a model with that of other models in the archive is also provided to further support depositors, journal editors and referees.
The reports will be sent to depositors of X-ray crystal structures upon completion of the curation process by wwPDB annotation staff. Later this year, a standalone web service will be released which can be used to generate validation reports prior to deposition. Please note that the reports and the online documentation are still undergoing active development. This means that there may be a few cases where a report cannot be generated (we endeavour to fix such cases as soon as we can), and that the content and layout of the reports is subject to change. However, the wwPDB partners feel that the benefits of providing the new reports vastly outweigh these issues. Early in 2014, validation reports for all X-ray crystal structures already in the PDB archive will be made publicly available as PDF files, together with XML files that contain exhaustive validation information for each entry (e.g., complete lists of outliers for every criterion). Validation pipelines for structures determined by NMR spectroscopy [3] or 3D cryo-electron microscopy [4] are under development and will be introduced at a later stage.
Further information, including sample X-ray validation reports, is provided. We welcome your feedback on the new validation reports. If you would like to send us your comments or questions, then please contact validation@mail.wwpdb.org
[1] Read RJ, Adams PD, Arendall WB, III, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH. (2011) A new generation of crystallographic validation tools for the Protein Data Bank.
Structure 19: 1395-1412. doi:
10.1016/j.str.2011.08.006
[2] Gore S., Velankar S. and Kleywegt G.J. Implementing an X-ray validation pipeline for the Protein Data Bank 2012,
Acta Cryst., D
68, 478-483.
doi:
10.1107/S0907444911050359
[3] Montelione GT, Nilges M, Bax A, Güntert P, Herrmann T, Richardson JS, Schwieters C, Vranken WF, Vuister GW, Wishart DS, Berman HM, Kleywegt GJ, Markley JL. (in press) Recommendations of the wwPDB NMR Structure Validation Task Force. Structure.
[4] Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schroder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL. (2012) Outcome of the first electron microscopy validation task force meeting.
Structure 20: 205-214.
doi: 10.1016/j.str.2011.08.006
07/18/2013
Upcoming Meetings: 3DSig, ACA, and ICSG
Get the latest news on the Common Deposition & Annotation System project and other initiatives at these events:
3Dsig:Structural Bioinformatics & Computational Biophysics satellite meeting (July 19-20; Berlin, Germany): Stephen Burley will present a keynote presentation and poster on wwPDB: Ensuring a freely accessible, singular archive of high quality macromolecular structure information.
ACA (American Crystallographic Association, July 20-24; Honolulu, HI): Helen Berman will present The wwPDB: Ensuring a single, uniform archive of high quality data during the session on Enabling Partnerships for Broader Crystallographic Data Accessibility. John Westbrook will present a poster and a tutorial in the Structure Validation session on The New wwPDB Deposition and Annotation System.
Demonstrations of the new system will be available at exhibit booth #101.
ICSG2013-SLS: At Structural Life Science/Seventh International Conference on Structural Genomics (July 29-August 1; Sapporo, Japan), Stephen Burley will present wwPDB: Ensuring a freely accessible, singular archive of high quality macromolecular structure information in a talk and a poster.
07/01/2013
Celebrating 10 Years of the wwPDB
July 1st 2013 marks the 10-year anniversary of the founding of the Worldwide Protein Data Bank (wwPDB), the international collaboration that manages the PDB archive (1).

2003
2004
2005
2006
2007
2008
2009
2010
2011
2012
2013
Click on the birthday cake for a slideshow of wwPDB milestones through the years.
From modest beginnings
Starting from just 7 protein crystal structures in 1971, the PDB archive has grown rapidly over the past 42 years. Last year alone, 9,972 new structures were deposited, more than in the first 25 years of the PDB combined. Today, the archive contains over 90,000 structures and at its current rate of growth will reach the 100,000 structure mark in 2014, the International Year of Crystallography.
On July 1st 2003, the way in which the PDB archive was managed was transformed by the founding of the Worldwide Protein Data Bank organization. From its inception, the PDB has been an international archive and the establishment of the wwPDB ensured that these valuable data will continue to be stored, managed and kept freely available for the benefit of scientists worldwide.
The wwPDB organization nowadays consists of four partners: Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) and BioMagResBank (BMRB) in the USA, Protein Data Bank in Europe (PDBe), and Protein Data Bank Japan (PDBj).
wwPDB activities
The wwPDB partner sites each act as deposition, processing and distribution centres for PDB data. They work together and in consultation with the wider community to de�ne deposition and annotation policies, �le formats and validation standards for structural data. This close collaboration between the member organizations is vital to guarantee that the global community of PDB users is provided with reliable and consistent data.
While working jointly on all aspects of data representation and processing, each partner site also offers independent tools and services that help make the wealth of data about biomacromolecular structure and function easily accessible to the user community.
wwPDB activities are overseen by an international advisory committee comprising of experts in X-ray crystallography, 3DEM, NMR, and bioinformatics.
Future Challenges
The increasing volume, diversity and complexity of biological data being deposited in the PDB and the emergence of hybrid techniques to obtain structural insights into biologically relevant molecules, complexes and molecular machines all present major challenges for the management and presentation of structural data.
To address these challenges, the wwPDB partners are jointly developing a software system that will allow deposition, validation and annotation of complex and diverse macromolecular structures along with the underlying experimental data using a single interface. This new system will go into full production at all the wwPDB deposition sites early in 2014 and will be able to handle depositions of structures of any size, determined using diffraction, NMR and/or EM methods.
Validation will be an integral part of the new deposition and annotation system. Assessment of coordinates, experimental data and associated meta data at the time of deposition is vital for improving the quality of the archive. In addition, it will help users with no or limited structural biology background to select the most appropriate structural models for their purposes.
Whatever new challenges the next 10 years will bring, the wwPDB will remain committed to maintain high standards of quality, integrity and consistency of the macromolecular structure archive and to make it freely available to an increasingly large, diverse and demanding global community of users.
(1) Announcing the worldwide Protein Data Bank. Berman H, Henrick K, Nakamura H. Nat. Struct. Biol. 10, 980 (2003) doi:10.1038/nsb1203-980
06/19/2013
10,000 NMR structure milestone reached
The wwPDB is excited to announce that the number of structures available in the PDB archive determined using Nuclear Magnetic Resonance (NMR) spectroscopy has passed the 10,000 mark.
NMR and the PDB
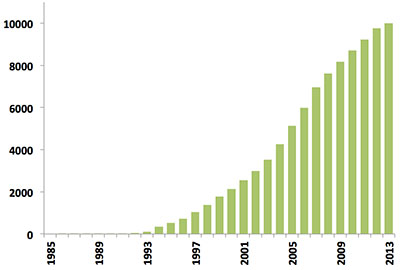
Since the first biomacromolecular NMR structure was archived in 1989, the number of NMR-derived structures in the PDB has grown steadily. Last year alone over 500 new NMR structures were deposited, more than in the first 10 years of NMR depositions combined (Figure 1). Today, NMR-derived structures account for more than 10% of the PDB archive which itself will reach the 100,000 structure mark in 2014.
Figure 1: Yearly growth of released structures in the PDB solved by NMR
A typical NMR structure in the PDB will consist of an ensemble of models. Nowadays, in addition to the structural ensemble, wwPDB requires the deposition of the assigned chemical shifts as well as the geometric restraints used in the structure determination and refinement. Chemical shifts and restraints are then shared with the BMRB archive, the main repository of experimental NMR data.
NMR structures in the PDB and associated experimental data are presented by the wwPDB partner sites through entry-specific pages. Additionally, a number of dedicated databases, tools and services further help make this wealth of NMR structural information accessible to the scientific community (Table 1).
Future Challenges
As the PDB archive continues to grow, the provision of robust validation tools is becoming even more important. The wwPDB partners are actively driving the development of new standards and software for the validation of structures determined by NMR spectroscopy. A wwPDB NMR Validation Task Force (VTF) has been convened to define standard validation criteria, representing community consensus. The VTF has provided its initial recommendations, which will be implemented in a wwPDB validation pipeline and applied to all depositions of NMR structures.
Table 1: Web resources for NMR structures exemplified using PDB entry 2LPZ.
Structure of a bacterial type III secretion needle from Salmonella typhimurium (PDB entry 2LPZ; BMRB entry 18276; Loquet et al., 2012, Nature, 486:276) determined using a hybrid solid-state NMR and electron microscopy (EM) approach.
06/12/2013
Small Angle Scattering Task Force
We are pleased to announce that the Report of the wwPDB Small-Angle Scattering Task Force, "Data Requirements for Biomolecular Modeling and the PDB Structure", has been published in the journal Structure (doi:10.1016/j.str.2013.04.020).
The first meeting of the Small Angle Scattering (SAS) Task Force (July 12-13, 2012) was sponsored by the wwPDB and held at the Center for Integrative Proteomics Research at Rutgers, The State University of New Jersey. The Task Force, chaired by Jill Trewhella, includes experts in SAS, crystallography, data archiving, and molecular modeling.
Recognizing the rapidly growing community of structural biology researchers that acquire and interpret SAS data in terms of increasingly sophisticated molecular models, the SAS Task Force made several recommendations. These include: development of a global repository for X-ray and neutron SAS data; creation of a standard dictionary of terms for data collection and for managing the SAS data repository; options for including SAS-derived shape and atomistic models along with specific information regarding the modeling protocol, uniqueness and uncertainty; development of criteria for assessment of data quality and accuracy. The Task Force also recommends that leaders from the various structural biology disciplines should jointly define what to archive in the PDB and what complementary archives might be needed, taking into account both scientific needs and funding.
This report by the wwPDB SAS Task Force follows recommendations recently published by wwPDB Validation Task Forces on X-ray and 3DEM. The report from the NMR Validation Task Force will be published shortly.
05/30/2013
Landmark HIV Capsid Structures Released in PDB

Two complete HIV-capsid structures, both of unprecedented size, are described in this week's issue of Nature
and released in the Protein Data Bank (PDB; www.wwpdb.org). This represents a significant advance in the field of structural biology and a milestone for the PDB.
PDB entries 3J3Q and 3J3Y are models based on cryo-electron microscopy data and use of a molecular dynamics flexible-fitting method. They contain 1356 and 1176 protein chains, respectively, and over two million atoms each. The HIV-1 capsid is the protein envelope that encloses and protects the RNA genome of the virus. An important subject of study, the full capsid has been a difficult target for structural characterization due to its extremely large size and morphological variability.
The wwPDB has anticipated structures of increasing size and complexity that exceed the limitations of the original PDB file format. These capsid structures have been curated following the recently announced wwPDB procedures for the deposition and release of large structures. Extremely large structures can now be deposited, annotated, and released as single files in PDBx/mmCIF and PDBML/XML formats.
The intact capsid structures and the cryo-electron tomography reconstruction from which they were generated can be downloaded from the PDB archive ftp sites in the USA, Europe and Japan as follows:
Reference
Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Gongpu Zhao, Juan R. Perilla, Ernest L. Yufenyuy, Xin Meng, Bo Chen, Jiying Ning, Jinwoo Ahn, Angela M. Gronenborn, Klaus Schulten, Christopher Aiken, & Peijun Zhang,
Nature 497, 643-646 (2013) DOI: 10.1038/nature12162
Related Entries
EMDB entry EMD-5639 is the cryo-electron tomography reconstruction from which 3J3Q and 3J3Y were generated; related entry 3J34, derived from an 8.6 Ångström reconstruction of a capsid hexameric subunit in a helical assembly (EMD-5582), was used in the construction of both 3J3Q and 3J3Y. These entries can be accessed and analysed through the websites of the wwPDB partners in Europe (pdbe.org), the USA (rcsb.org) and Japan (pdbj.org).
05/22/2013
Deposition and Release of PDB Entries Containing Large Structures
In order to meet the challenges of ever greater numbers of PDB depositions, involving ever larger and more complex structures, often determined using multiple methods, the Worldwide Protein Data Bank partners (wwPDB; www.wwpdb.org/) are developing a completely new system for deposition and annotation of PDB entries. This new system will go into full production at all the wwPDB deposition sites early in 2014 and will then be able to handle depositions of structures of any size, determined using diffraction, NMR and/or EM methods. Large structures will also be processed and released intact so that "split entries" become a thing of the past.
Currently, submitting a large structure that exceeds PDB format restrictions (e.g., more than 62 chains or more than 99,999 atoms) requires the depositor to split the large structure into multiple PDB files. The new wwPDB deposition and annotation system will be able to handle such structures if they are provided in a single file in PDBx/mmCIF format. The PDBx/mmCIF (http://mmcif.pdb.org/) format does not suffer the restrictions of the PDB format and is capable of representing large structures in a single file.
With the new deposition system, large structures that are deposited as a single PDBx/mmCIF file will also be processed and released into the PDB ftp archive as a single PDBx/mmCIF file (along with a companion PDBML format file). Since the coordinate records in such structures cannot be validly represented by the PDB format, only an abbreviated PDB formatted file, containing authorship and citation details, will be provided in the ftp archive. In addition, a web service will be provided by the wwPDB to translate the coordinate records and limited metadata into a collection of limited, "best-effort" PDB-format files. (Structures that do not exceed the limitations of the PDB format will continue to be provided as PDB files in the archive for the foreseeable future.)
The wwPDB has convened a Working Group for PDBx/mmCIF Data Deposition (PDBx/mmCIF Working Group) that includes representatives from the major X-ray structure-determination packages, and is chaired by Paul Adams. In order to ease the transition from PDB to PDBx/mmCIF, the Working Group has made recommendations about essential extensions required for large structures, including:
- Atom serial numbers will run from 1 to the number of deposited atoms (with no field-width restrictions);
- Chain identifiers of up to 4 characters will be allowed;
- Cartesian coordinates will have field width as large as required (and maintain 3 decimal places precision);
- Isotropic B-factors and occupancies will have 3 decimal places precision.
PDBx/mmCIF files suitable for deposition can now be created with recent versions of the CCP4 (REFMAC 5.8) and Phenix (1.8.2) software packages. Both packages support the above extensions for large structures.
Although the new wwPDB deposition and annotation system will not go into full production until early 2014, limited external testing will commence in August 2013. We invite depositors of large structures (that currently break the PDB format restrictions and necessitate splitting of entries) to contact info@wwpdb.org to gain access to the new system as beta testers.
Prior to the release of the new deposition system in 2014, depositors needing to submit large entries as a single PDBx/mmCIF data file should contact info@wwpdb.org. The wwPDB will arrange to accept and process the large entries intact and provide a special download location on the wwPDB ftp site for the large entry (ftp://ftp.wwpdb.org/pub/pdb/data/large_structures). In order not to break any current software systems, wwPDB curators will "split" such an entry into a collection of PDB-format files that will be distributed on the wwPDB ftp site following current release and formatting conventions. In 2014, all legacy split entries will be "reunited" and released intact (in PDBx/mmCIF format only) by the wwPDB.
Users with questions about the new deposition system or the procedures for handling large structures should contact info@wwpdb.org
04/02/2013
The Biologically Interesting Molecule Reference Dictionary (BIRD) for Peptide-like Antibiotic and Inhibitor Molecules
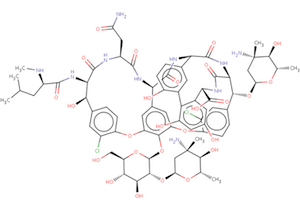
The wwPDB's Biologically Interesting molecule Reference Dictionary (BIRD) describes antibiotics, peptide inhibitors, and other complex biological ligands. To help define and represent these biologically interesting molecules, BIRD contains chemical descriptions, sequence and linkage information, and functional and classification information as taken from the core structures and from external resources.
All PDB entries containing these molecules have been annotated using this dictionary, with corresponding BIRD ID code contained only in the PDBx-formatted file. The use of BIRD will greatly improve the consistency of peptide-like antibiotic and inhibitor molecules in the PDB.
BIRD is available on the wwPDB FTP server adjacent to the Chemical Component Dictionary at ftp://ftp.wwpdb.org/pub/pdb/data/bird/prd/. It is updated as new entries are added to the PDB archive. An overview of BIRD, along with definition details, is available online.
These data reflect the wwPDB's continuing commitment to providing accurate and detailed data to users worldwide
03/06/2013
Time-stamped Copies of the PDB Archive
A snapshot of the PDB archive (ftp.wwpdb.org) as of January 1, 2013 has been added to ftp://snapshots.wwpdb.org/. Snapshots have been archived annually since January 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20130101 includes the 87,090 experimentally-determined coordinate files and related experimental data that were available at that time. Coordinate data are available in PDB, mmCIF, and XML formats. The date and time stamp of each file indicates the last time the file was modified.
The script at ftp://snapshots.wwpdb.org/rsyncSnapshots.sh may be used to make a local copy of a snapshot or sections of the snapshot.