wwPDB 2021 News
Contents
12/21/2021
Paper Published in Glycobiology
 Glycobiology
GlycobiologyCarbohydrate molecules present in more than 14,000 PDB structures were reviewed and remediated to conform to a new standardized format to facilitate broader usage of the resource by the glycoscience community and researchers studying glycoproteins.
Modernized uniform representation of carbohydrate molecules in the Protein Data Bank
Chenghua Shao, Zukang Feng, John D Westbrook, Ezra Peisach, John Berrisford, Yasuyo Ikegawa, Genji Kurisu, Sameer Velankar, Stephen K Burley, Jasmine Y Young
(2021) Glycobiology 31: 1204–1218, doi: 10.1093/glycob/cwab039
Additional documentation about the carbohydrate remediation project is also available.
12/17/2021
Biocurator Milestone: >10,000 Depositions Processed
Congratulations to the wwPDB biocurator Dr. Brian Hudson on processing over 10,000 PDB depositions! Dr. Hudson received his PhD in Chemistry from the California Institute of Technology and has an expertise in X-ray crystallography and cryo-electron microscopy. He has joined PDB in 2010 and has established himself as a highly qualified professional with deep understanding of scientific data and various experimental techniques and dedication to exceptional quality data curation. He was one of the EM data curation pioneers who curated over 1000 of EM map entries before the OneDep system was established. We congratulate Dr. Hudson with this exciting accomplishment and look forward to his further career success!
12/14/2021
Deposition of Half-maps for Certain EM Entries to Become Mandatory
From February 25, 2022, deposition of half-maps for single-particle, single-particle-based helical, and sub-tomogram averaging reconstructions to the EM Data Bank (EMDB) will become mandatory. This change is in response to a long-standing community request to the wwPDB EMDB Core Archive and was also a recommendation from the 2020 wwPDB single-particle cryo-EM data-management workshop (white paper in preparation). Several recommendations from this workshop have already been implemented in the wwPDB OneDep system. These include improvements to wwPDB validation reports and enhancements for capturing metadata via the deposition interface.
Mandatory half-maps must be unfiltered, unmasked, unsharpened, and positioned in the same coordinate-space and orientation as the primary map such that they superimpose. The availability of half-maps will contribute to improved validation of EM structures as reflected in the wwPDB validation reports.
wwPDB strongly urges developers of cryo-EM processing software for the affected modalities to implement support for output of such half-maps (if this is not already available).
Any queries about this policy change can be directed to info@wwpdb.org.
12/08/2021
Watch Workshops on Open-Source Tools for Chemistry
 The RSC Chemical Information and Computer Applications Group celebrated PDB50 in November 2021
The RSC Chemical Information and Computer Applications Group celebrated PDB50 in November 2021As part of their series of Workshops on Open-Source Tools for Chemistry, the Chemical Information and Computer Applications Group of the Royal Society of Chemistry hosted two PDB50 celebrations on November 16 and 18, 2021. Videos of these presentations are available online.
Protein Data Bank at 50: Accessing, Understanding, and Assessing PDB Data
- History of the Protein Data Bank (PDB) and the Worldwide PDB, Stephen K. Burley, RCSB Protein Data Bank (RCSB PDB)
- PDBx/mmCIF Data Standard and PDB Data Deposition with OneDep, Ezra Peisach and Jasmine Young, RCSB PDB
- Small Molecules in the PDB, Chenghua Shao, RCSB PDB
- OneDep and Small Molecules Tutorial, Chenghua Shao and Ezra Peisach, RCSB PDB
- wwPDB Validation: Assessing the Quality of PDB structures, John Berrisford, Protein Data Bank in Europe (PDBe)
- How to Interpret the Quality of a PDB Structure using the wwPDB validation report, David Armstrong, John Berrisford, Jack Turner, PDBe
- 3D Visualization of PDB Data with the Mol* viewer James Tolchard, PDBe
- Impact of AI and Future of PDB Data: Next Generation PDB Archive, Sameer Velankar (PDBe)
- Round Table Discussion, Sameer Velankar (PDBe) and Stephen K. Burley (RCSB PDB)
After these workshops, attendees should be able to:
- Access and appropriately use the wwPDB OneDep system for macromolecular structure depositions
- Understand how small molecule data in the PDB is organized and how to access it
- Evaluate the quality and accuracy of a PDB structure using the wwPDB validation report
- Understand how to use the Mol* molecular viewer to visualize PDB structures
- Appreciate wwPDB plans for establishing a Next Generation PDB Archive
11/23/2021
Watch Presentations from the October 6 PDB50 Celebration
The Biophysical Society hosted a virtual symposium on October 6, 2021, highlighting some of the high-impact applications of protein structural data, with a particular focus on the areas of structure prediction and membrane protein biophysics.
The recorded presentations from that day are available from the BPS Video Library
Session I. Enabling Understanding of Protein Structure, Function, and Design
- Helen M. Berman, Rutgers - The State University of New Jersey and RCSB PDB
- John Jumper, DeepMind, Inc, United Kingdom
- Ruth Nussinov, NIH, USA and Tel Aviv University, Israel
- Christine Orengo, University College London, United Kingdom
- David Baker, University of Washington, USA
Session II. Molecular Biophysics of Membrane Proteins
- Stephen K. Burley, Rutgers - The State University of New Jersey and RCSB PDB
- Jue Chen, Rockefeller University and HHMI, USA
- Nieng Yan, Princeton University, USA
- Linda Columbus, University of Virginia, USA
- Rod MacKinnon, Rockefeller University and HHMI, USA
Organizers
- Helen M. Berman, Rutgers - The State University of New Jersey and RCSB PDB, USA
- Stephen K. Burley, Rutgers - The State University of New Jersey and RCSB PDB, USA
- Gaetano T. Montelione, Rensselaer Polytechnic Institute, USA
11/02/2021
November Workshops on Open-Source Tools for Chemistry
As part of their series of Workshops on Open-Source Tools for Chemistry, the Chemical Information and Computer Applications Group of the Royal Society of Chemistry will be hosting two free virtual events in honor of PDB50.
Protein Data Bank at 50: Accessing, Understanding, and Assessing PDB Data
Day 1: Tuesday November 16, 2021 | 3-5pm GMT
- History of the Protein Data Bank (PDB) and the Worldwide PDB, Stephen K. Burley, RCSB Protein Data Bank (RCSB PDB)
- PDBx/mmCIF Data Standard and PDB Data Deposition with OneDep, Ezra Peisach and Jasmine Young, RCSB PDB
- Small Molecules in the PDB, Chenghua Shao, RCSB PDB
- OneDep and Small Molecules Tutorial, Chenghua Shao and Ezra Peisach, RCSB PDB
Day 2: Thursday November 18 2021 3-5pm GMT
- wwPDB Validation: Assessing the Quality of PDB structures, John Berrisford, Protein Data Bank in Europe (PDBe)
- How to Interpret the Quality of a PDB Structure using the wwPDB validation report, David Armstrong, John Berrisford, Jack Turner, PDBe
- 3D Visualization of PDB Data with the Mol* viewer James Tolchard, PDBe
- Impact of AI and Future of PDB Data: Next Generation PDB Archive, Sameer Velankar (PDBe)
- Round Table Discussion, Sameer Velankar (PDBe) and Stephen K. Burley (RCSB PDB)
After these workshops, attendees should be able to:
- Access and appropriately use the wwPDB OneDep system for macromolecular structure depositions
- Understand how small molecule data in the PDB is organized and how to access it
- Evaluate the quality and accuracy of a PDB structure using the wwPDB validation report
- Understand how to use the Mol* molecular viewer to visualize PDB structures
- Appreciate wwPDB plans for establishing a Next Generation PDB Archive
Please register to attend these Open-Source Tools for Chemistry Workshops.
10/27/2021
Obituary for John Westbrook
 John Westbrook at the 2017 Congress and General Assembly of the International Union of Crystallography in Hyderabad, India
John Westbrook at the 2017 Congress and General Assembly of the International Union of Crystallography in Hyderabad, IndiaJohn D. Westbrook Jr. (1957-2021), Research Professor at Rutgers University and Data & Software Architect Lead for the RCSB PDB, passed away on October 18, 2021.
He was incredibly beloved and respected by his colleagues at Rutgers and throughout the world, known for his dry wit and endless enthusiasm for thinking about all aspects of data and data management.
John had a long and highly successful career developing ontologies, tools, and infrastructure in data acquisition, validation, standardization, and mining in the structural biology and life science domains. His work established the PDBx/mmCIF data dictionary and format as the foundation of the modern Protein Data Bank (PDB) archive (wwPDB.org).
More than twenty-five years ago, while still a graduate student, John recognized the importance of a well-defined data model for ensuring delivery of high quality and reliable structural information to data users. He was the principal architect of the mmCIF data representation for biological macromolecular data. Based on a simple, context-free grammar (without column width constraints), data are presented in either key-value or tabular form. All relationships between common data items (e.g., atom and residue identifiers) are explicitly documented within the PDBx Exchange Dictionary (mmcif.wwpdb.org). Use of the PDBx/mmCIF format enables software applications to evaluate and validate referential integrity within any PDB entry. A key strength of the mmCIF technology is the extensibility afforded by its rich collection of software-accessible metadata.
The current PDBx/mmCIF dictionary contains more than 6,200 definitions relating to experiments involved in macromolecular structure determination and descriptions of the structures themselves. The first implementation of this schema was used for the Nucleic Acid Database, a data resource of nucleic acid-containing X-ray crystallographic structures. Today, this dictionary underpins all data management of the PDB. Since 2014, it has served as the Master Format for the PDB archive. It also forms the basis of the Chemical Component Dictionary (wwpdb.org/data/ccd), which is used to maintain and distribute small molecule chemical reference data in the PDB.
In 2011, the Worldwide Protein Data Bank (wwPDB) PDBx/mmCIF Working Group was established to enable direct use of PDBx/mmCIF format files within major macromolecular crystallography software tools and to provide recommendations on format extensions required for deposition of larger macromolecule structures to the PDB. This was a key step in the evolution of the PDB archive, which enabled studies of macromolecular machines, such as the ribosome, as single PDB structures (instead of split entries with atomic coordinates distributed among different entry files). In 2019, mandatory submission of PDBx/mmCIF format files for deposition was announced (Adams et al. Acta Crystallographica D75, 451-454).
To ensure the success of the PDBx/mmCIF dictionary and format, John worked with a wide range of community experts to extend the framework to encompass descriptions of macromolecular X-ray crystallographic experiments, 3D cryo-electron microscopy experiments, NMR spectroscopy experiments, protein and nucleic acid structural features, diffraction image data, and protein production and crystallization protocols. Most recently, these efforts have been focused on developing compatible data representations for X-ray free electron (XFEL) methods, and for integrative or hybrid methods (I/HM). I/HM structures, currently stored in the prototype PDB-Dev archive (pdb-dev.wwpdb.org), presented new challenges for data exchange among rapidly evolving and heterogeneous experimental repositories. Proper management of I/HM structures in PDB-Dev also required extension of the PDBx/mmCIF data dictionary to include coarse-grained or multiscale models, which will be essential for studying macromolecular structures in situ using cryo-electron tomography and other bioimaging methods.
John contributed broadly to community data standards enabling interoperation and data integration within the biology and structural biology domains. His efforts have included (i) describing the increasing molecular complexity of macromolecular structure data, (ii) representing new experimental methodologies, including I/HM techniques, and (iii) expanding the biological context required to facilitate broader integration with a spectrum of biomedical resources. John’s work has been central to connecting crystallographic and related structural data for biological macromolecules to key resources across scientific disciplines. His efforts have been described in more than 120 peer-reviewed publications, one of which has been cited more than 21,000 times according to the Web of Science (Berman et al. Nucleic Acids Research 28, 235-242). Eight of his most influential published papers have appeared in the International Tables of Crystallography.
John has also done yeoman service to the crystallographic community over many years and was recognized with the inaugural Biocuration Career Award from the International Society for Biocuration in 2016.
For the International Union of Crystallography, John served on the Commission for Maintenance of CIF Standard (COMCIFS), the Working Group on Data Diffraction Deposition (DDDWG), and the Committee on Data (CommDat). He also served as an Associate Editor for Acta Crystallographica Section F.
John was a long-standing member of the American Crystallographic Association, and served on the Data, Standards & Computing Committee. He also served on the Metadata Interest Group for the Research Data Alliance.
John is survived by his wife, Bonnie J. Wagner-Westbrook, Ed.D. and his devoted Mother-in-Law, Joan N. Wagner of Clinton Twp., NJ; many cousins including Chandler Turner (of Portsmouth, VA), Ann (Turner) Heyes (of Tasmania, Australia) and Louise (Turner) Brown (of Oakland CA).
Visitation will take place on Saturday, November 6, 2021 from 2-4pm with Memorial Service at 4pm. All at Scarponi-Bright Funeral Home, 26 Main Street, Lebanon, NJ. Interment will be private.
Memorials can be made to Capicats or an organization of choice in his honor.
Additional information is available at Scarponi-Bright.
Obituary
John D. Westbrook Jr (1957–2021) Acta Cryst (2021) D77: 1475-1476 doi: 10.1107/S2059798321011402
Dedications
- RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D (2022) Protein Science 31: 187-208 doi: 10.1002/pro.4213
- Collecting Experiments. Making Big Data Biology. Helliwell, J. R. (2022). J. Appl. Cryst. 55: 211-214 doi: 10.1107/S1600576721012140
- PDBx/mmCIF Ecosystem: Foundational Semantic Tools for Structural Biology (2022) Journal of Molecular Biology 434: 167599 doi: 10.1016/j.jmb.2022.167599
- RCSB Protein Data Bank: improved annotation, search and visualization of membrane protein structures archived in the PDB (2022) Bioinformatics 38: 1452-1454 doi: 10.1093/bioinformatics/btab813
10/19/2021
PDB Turns 50
The PDB was announced on October 20, 1971 in Crystallography: Protein Data Bank Nature New Biology 233: 223 (1971) doi: 10.1038/newbio233223b0.
Today, the PDB archive contains >180,000 structures of proteins, nucleic acids, and complex assemblies that helps students and researchers understand all aspects of biomedicine and agriculture, from protein synthesis to health and disease. It is managed by the Worldwide PDB (wwPDB) organization that ensures that the PDB is freely and publicly available to the global community.
The wwPDB has been celebrating this golden anniversary with symposia and events throughout 2021.
Consider supporting 50 years of PDB's spirit of openness, cooperation, and education with a donation to the wwPDB Foundation. The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB50 events, workshops, and educational publications.
The Foundation is chartered as a 501(c)(3) entity exclusively for scientific, literary, charitable, and educational purposes.
10/19/2021
PDB50 Anniversary Symposium in Asia
10/12/2021
Biocurator Milestone: >10,000 Depositions Processed
Congratulations to biocurators Dr. Sutapa Ghosh and Dr. Monica Sekharan on processing over 10,000 PDB depositions. They are the second and third biocurators to reach this milestone. Yumiko Kengaku reached this milestone in April 2021.
Dr. Ghosh received her PhD in structural biology from the University of Calcutta and joined PDB after working in industry in structure-based drug design. Dr. Sekharan received her PhD in Biological Chemistry from the University of Washington with expertise in NMR spectroscopy. During their 15 year career at the PDB, many depositors trusted their professional skills in accurate and comprehensive data analysis and representation. Their deep scientific knowledge, profound data curation expertise and commitment to excellence contributed to the high quality data archive for the benefit of the scientific community. We congratulate Drs. Ghosh and Sekharan with this exciting accomplishment and look forward to their future successes.
 RCSB PDB Biocurators Dr. Sutapa Ghosh and Dr. Monica Sekharan
RCSB PDB Biocurators Dr. Sutapa Ghosh and Dr. Monica Sekharan
09/24/2021
PDBx/mmCIF data files to include PI information
 PI name, email, and ORCiD ID will be publicly available in PDBx/mmCIF data files starting September 24, 2021
PI name, email, and ORCiD ID will be publicly available in PDBx/mmCIF data files starting September 24, 2021wwPDB continues to support research, education, and drug discovery worldwide. Open access to PDB data has helped researchers in structure-guided discovery and development of anti-coronavirus drugs, vaccines and neutralizing antibodies. When researchers analyze existing PDB structures, such as working on a similar structure, they may often need additional information impossible to retrieve from the PDB entry file alone. In particular, it is not possible to obtain a point of contact in cases where there is no associated primary publication for an entry.
Following a recommendation from the IUCr Commission on Biological Macromolecules and the IUCr Committee on Data, wwPDB will make public the PI name, email address, and ORCiD ID for initial PDB depositions or re-submissions made, starting September 24, 2021. This will enable contact with the authors of every released PDB structure as of that date. This release will also align the PDB with the standard practices of providing corresponding author information by scientific journals
The dated acceptance of these PDB Terms and Conditions described above will be captured within the OneDep system. The responsible depositor who creates the deposition should make entry PI(s) aware of the policy change to include PI name, email address, and ORCiD in public PDBx/mmCIF files.
09/21/2021
Register for an October 6 PDB50 Celebration
The Biophysical Society will host a virtual symposium on October 6, 2021, highlighting some of the high-impact applications of protein structural data, with a particular focus on the areas of structure prediction and membrane protein biophysics.
Registration is free, however space is limited. Registration deadline is October 4.
Session I. Enabling Understanding of Protein Structure, Function, and Design
- Helen M. Berman, Rutgers - The State University of New Jersey and RCSB PDB
- John Jumper, DeepMind, Inc, United Kingdom
- Ruth Nussinov, NIH, USA and Tel Aviv University, Israel
- Christine Orengo, University College London, United Kingdom
- David Baker, University of Washington, USA
Session II. Molecular Biophysics of Membrane Proteins
- Stephen K. Burley, Rutgers - The State University of New Jersey and RCSB PDB
- Jue Chen, Rockefeller University and HHMI, USA
- Nieng Yan, Princeton University, USA
- Linda Columbus, University of Virginia, USA
- Rod MacKinnon, Rockefeller University and HHMI, USA
Organizers
- Helen M. Berman, Rutgers - The State University of New Jersey and RCSB PDB, USA
- Stephen K. Burley, Rutgers - The State University of New Jersey and RCSB PDB, USA
- Gaetano T. Montelione, Rensselaer Polytechnic Institute, USA
09/14/2021
Improved Access to Chemical Component Definitions and Archive Inventories
 Individual Chemical Component Dictionary entries and new archive inventory lists are now available for download
Individual Chemical Component Dictionary entries and new archive inventory lists are now available for downloadImproved access to small molecule definitions
Individual Chemical Component Dictionary (CCD) and Biologically Interest molecule Reference Dictionary (BIRD) definitions are now accessible in a new FTP tree in the PDB archive. In response to user requests, these individual CCD and BIRD entry files can be found at /pdb/refdata/chem_comp/ and /pdb/refdata/bird/, respectively with last character hash as sub-directory.
For example:
- /pdb/refdata/chem_comp/C/D8C/D8C.cif
- /pdb/refdata/bird/prd/8/PRD_001068.cif
Improved access to information about PDB archive holdings
New inventory data files offer a quick overview of data in the archive. These files are in the extensible JSON format, and can be found under the new /pdb/holdings/ FTP tree.
The inventory lists provided include:
- all_removed_entries.json.gz: list of removed PDB entries (obsolete, models) with entry authors, entry title, release date, obsolete date , and superseding PDB ID, if any.
- current_file_holdings.json.gz: List of released PDB entries and file types present for each entry in the PDB Core Archive (e.g., coordinate data, experimental data, validation report, ...)
- obsolete_structures_last_modified_dates.json.gz: List of obsolete PDB entries with last time of PDBx/mmCIF file modification
- refdata_id_list.json.gz: List of released chemical reference entries, content types (e.g., Chemical Component, BIRD), and last time of reference file modification
- released_structures_last_modified_dates.json.gz: List of released PDB entries with last time of PDBx/mmCIF file modification
- unreleased_entries.json.gz: List of on-hold PDB entries, entry status, deposition date, and sequence pre-release information
The inventory (index) files historically provided in /pdb/derived_data/ will continue to be updated for the time being; they will eventually be removed from the PDB archive. Users are encouraged to utilize these new inventory files.
09/09/2021
Bringing Molecular Structure to Life
EMBL will host a virtual symposium on October 20-22, 2021 celebrating 50 years of the PDB.
Registration deadline is September 29.
Session Topics
- Structural biology and applications in health and the environment
- RNA/DNA molecular machines
- The next 50 years: Genomics meets structural biology
- Latest advances
- The next 50 years: Future perspectives (part 1)
- The next 50 years: Future perspectives (part 2)
Speakers
- Bissan Al-Lazikani, The Institute of Cancer Research, UK
- Cheryl H. Arrowsmith, University Health Network, Canada
- M. Madan Babu, St. Jude Children‘s Research Hospital, USA
- Drew Berry, Walter and Eliza Hall Institute of Medical Research, Australia
- Wah Chiu, Stanford University, USA
- Patrick Cramer, Max Planck Institute for Biophysical Chemistry, Germany
- Petra Fromme, Arizona State University, USA
- Donald Hilvert, ETH Zurich, Switzerland
- Martin Jinek, University of Zurich, Switzerland
- John Jumper, DeepMind, UK
- Julia Mahamid, EMBL Heidelberg, Germany
- Christine Orengo, University College London, UK
- Lori A. Passmore, MRC Laboratory of Molecular Biology, UK
- Jane Shelby Richardson, Duke University School of Medicine, USA
- David Stuart, University of Oxford, UK
- Nicolas Thomä, Friedrich Miescher Institute for Biomedical Research, Switzerland
- Janet Thornton, EMBL-EBI Hinxton, UK
- Sameer Velankar, EMBL-EBI Hinxton, UK
Panel Chair
- Peter Rosenthal, The Francis Crick Institute, UK
Organizers
- Stephen Cusack, EMBL Grenoble, France
- Gerard Kleywegt, EMBL-EBI, UK
- Christoph Mueller, EMBL Heidelberg, Germany
- Christine Orengo, University College London, UK
- Janet Thornton, EMBL-EBI, UK
- Sameer Valenkar, EMBL-EBI, UK
- Matthias Wilmanns, EMBL Hamburg, Germany
08/17/2021
PDB50 at ACS August 25
 The Fall 2021 ACS meeting will be hybrid.
The Fall 2021 ACS meeting will be hybrid.Celebrate PDB50 at the Fall 2021 ACS Meeting with a session on Understanding Enzyme Function in 3D: Celebrating 50 Years of the Protein Data Bank.
All times shown are are listed in Eastern Daylight Time (EDT) on Wednesday August 25, 2021.
2:00 Introductory Remarks
2:05 The winding road from G-quadruplexes to telomerase, Juli Feigon (UCLA)
2:45 Enhanced exploration of small-molecule ligands bound to proteins and nucleic acids, Stephen K. Burley (Rutgers University and UCSD)
3:10 Mechanistic insights into the cleavage and polyadenylation machinery, Lori Passmore (University of Cambridge)
3:35 Vive la difference! The synergies and differences between the PDB and the CSD, Jason Cole (CCDC)
4:00 Break
4:30 Beyond static snapshots of protein structure: The role of dynamics in function, George Phillips Jr. (Rice University)
5:10 Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the COVID-19 pandemic, Sagar Khare (Rutgers)
5:35 Cracking the phosphatase code: Holoenzyme formation, regulatory protein binding and susbtrate dephosphorylation by the phosphoprotein phosphatase family, Rebecca Page (Brown)
6:00 Small molecules targeting COVID-19 in an evolving landscape of publishing and peer review, James Fraser (UCSF)
6:30 Break
7:00 Watching metalloenzymes at work, Amie Boal (Penn State)
7:25 Time travel to the past and future – evolution of energy landscapes for enzymes catalysis, Dorothee Kern (Brandeis)
7:50 Structure, mechanism, and inhibition of class IIb histone deacetylases, David Christianson (Harvard)
8:15 Truth sometimes triumphs: The history of structural enzymology, Gregory Petsko (Brandeis)
8:55 Closing Remarks
Visit ACS for registration information.
This symposium was organized by Carmen Nitsche (CCDC), and Steven C. Almo (Albert Einstein College of Medicine), and Stephen K. Burley (RCSB PDB).
08/06/2021
wwPDB to switch to version 3 of the EMDB data model
From February 9, 2022, the wwPDB EMDB Core Archive will exclusively support version 3 of its data model and retire version 1.9.6 header files from the archive. The switch will involve several changes regarding file provision by the archive and this article outlines these changes.
Since the inception of OneDep in 2015, the EMDB Core Archive (EMBL-EBI: https://ftp.ebi.ac.uk/pub/databases/emdb/, PDBj: https://ftp.pdbj.org/pub/emdb/, wwPDB mirror site: https://ftp.wwpdb.org/pub/emdb/) has maintained two versions of its data model in parallel and also two versions of the header file for each entry. Currently, the official EMDB data model is version 1.9.6, while version 3, which facilitates a richer representation of the metadata about EMDB entries, was introduced in 2015.
Version 3 has now been finalized, and EMDB will therefore change its official data model version from v1.9.6 to v3. This will involve three changes to the EMDB Core Archive as of February 9, 2022:
- The current EMDB data model, defined in an XSD schema in a file named emdb.xsd and located in /doc/XML-schemas/emdb-schemas/current/, will change to the latest version of the v3 data model (at present v3.0.2.6).
- The official header file for each entry in the EMDB Core Archive, an XML file named emd-xxxxx.xml, will become a v3 XML file that follows the structure of the latest version of the v3 data model.
- The EMDB Core Archive will cease provision of v1.9.6 header files for all entries.
Currently, for an entry EMD-xxxxx in the EMDB core archive located at /structures/EMD-xxxxx/header/, the following header files are provided:
- Official header file: emd-xxxxx.xml
- Header file for data model v3: emd-xxxxx-v30.xml
- Header file for data model v1.9.6: emd-xxxxx-v19.xml
where emd-xxxxx.xml at present is a copy of the file emd-xxxxx-v19.xml, adhering to v1.9.6 of the EMDB data model.
From 9th February 2022 onward, for any entry, the following header files will be provided:
- Official header file: emd-xxxxx.xml
- Header file for data model v3: emd-xxxxx-v30.xml
where emd-xxxxx.xml will be a copy of emd-xxxxx-v30.xml, supported by v3 of the EMDB data model.
For any further information please email info@wwpdb.org.
08/01/2021
Extended PDB IDs and PDB DOIs now available in PDBx/mmCIF files
 Journals, PDB users, and software developers should review code and begin to prepare for the change in format of PDB IDs and inclusion of PDB DOIs in PDBx/mmCIF files
Journals, PDB users, and software developers should review code and begin to prepare for the change in format of PDB IDs and inclusion of PDB DOIs in PDBx/mmCIF fileswwPDB, in collaboration with the PDBx/mmCIF Working Group, has set plans to extend the length of ID codes for PDB and Chemical Component Dictionary (CCD) ID entries in the future. These extended formats are not supported by the legacy PDB file format.
As announced previously, wwPDB has extended PDB ID length to eight characters prefixed by ‘PDB’, e.g., pdb_00001abc.
Each PDB ID is issued a corresponding Digital Object Identifier (DOI), often required for manuscript submission to journals and described in publications by the structure authors.
To help depositors provide information to journals, OneDep now displays the PDB ID and DOI on the submission confirmation page.
The extended PDB IDs and corresponding PDB DOIs, along with existing four character PDB IDs, are now included in the PDBx/mmCIF formatted files. Initially, this will only be available for updated and newly-released PDB entries, with an archive-wide update at a later date.
The additional accessions will be provided in the _database_2 PDBx/mmCIF category.
For example, PDB entry 1ABC will have the extended PDB ID (pdb_00001abc) and the corresponding PDB DOI (10.2210/pdb1abc/pdb).
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB 1abc pdb_00001abc 10.2210/pdb1abc/pdb
WWPDB D_1xxxxxxxxx ? ?
Once all available four-character PDB IDs have been consumed, newly-deposited PDB entries will only be issued extended PDB ID codes. These entries will only be distributed in PDBx/mmCIF format.
wwPDB asks journals, users, and software developers to review code and remove related limitations.
07/26/2021
PDB50: Function Follows Form
The 2021 ACA Meeting Transactions Symposium Function Follows Form: Celebrating the 50th Anniversary of the Protein Data Bank celebrates this golden anniversary.
Friday July 30 Speakers
- Cynthia Wolberger - Johns Hopkins, Baltimore, MD
- Mike Martynowycz - HHMI/UCLA
- John Rubinstein - Sick Kid’s Hospital, Toronto, Canada
- Squire J. Booker - Penn State, State College, PA
- Rafael M. Couñago- SGC/UNICAMP, Brazil
- Erica Ollman Saphire - La Jolla Institute for Immunology, La Jolla, CA
Saturday July 31 Speakers
- Wayne A. Hendrickson - Columbia, New York, NY
- Wladek Minor - University of Virginia
- Chris Sander - Harvard Medical School, Boston, MA
- Eva Nogales - UC Berkeley/HHMI, Berkeley, CA
- Andrej Sali - RCSB PDB/UCSF, San Francisco, CA
Each day will end with a Panel Discussion: Leaning In – PDB in the Next 50 Years.
07/26/2021
PDBx/mmCIF data files to include PI information
 PI name, email, and ORCiD ID will be publicly available in PDBx/mmCIF data files starting September 24, 2021
PI name, email, and ORCiD ID will be publicly available in PDBx/mmCIF data files starting September 24, 2021wwPDB continues to support research, education, and drug discovery worldwide. Open access to PDB data has helped researchers in structure-guided discovery and development of anti-coronavirus drugs, vaccines and neutralizing antibodies. When researchers analyze existing PDB structures, such as working on a similar structure, they may often need additional information impossible to retrieve from the PDB entry file alone. In particular, it is not possible to obtain a point of contact in cases where there is no associated primary publication for an entry.
Following a recommendation from the IUCr Commission on Biological Macromolecules and the IUCr Committee on Data, wwPDB will make public the PI name, email address, and ORCiD ID for initial PDB depositions or re-submissions made, starting September 24, 2021. This will enable contact with the authors of every released PDB structure as of that date. This release will also align the PDB with the standard practices of providing corresponding author information by scientific journals
The dated acceptance of these PDB Terms and Conditions described above will be captured within the OneDep system. The responsible depositor who creates the deposition should make entry PI(s) aware of the policy change to include PI name, email address, and ORCiD in public PDBx/mmCIF files.
07/06/2021
Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures
An article describing updates and improvements to Mol* is highlighted on the cover of the 2021 Nucleic Acids Research Web Server Issue. As the primary 3D structure viewer used by PDBe and RCSB PDB, it enables 3D exploration of macromolecular coordinate and experimental data directly within the browser window. The project is an open collaboration started by PDBe, RCSB PDB, CEITEC, and welcomes new contributors.
Two outstanding students, Áron Samuel Kovács and Sukolsak Sakshuwong, contributed new functionality to Mol*. Thanks to their work, Mol* now has greatly improved 3D rendering capabilities and can also export molecular scenes as 3D object files for use in external rendering programs. These features can currently be previewed on molstar.org before they are made available at PDBe and RCSB PDB.
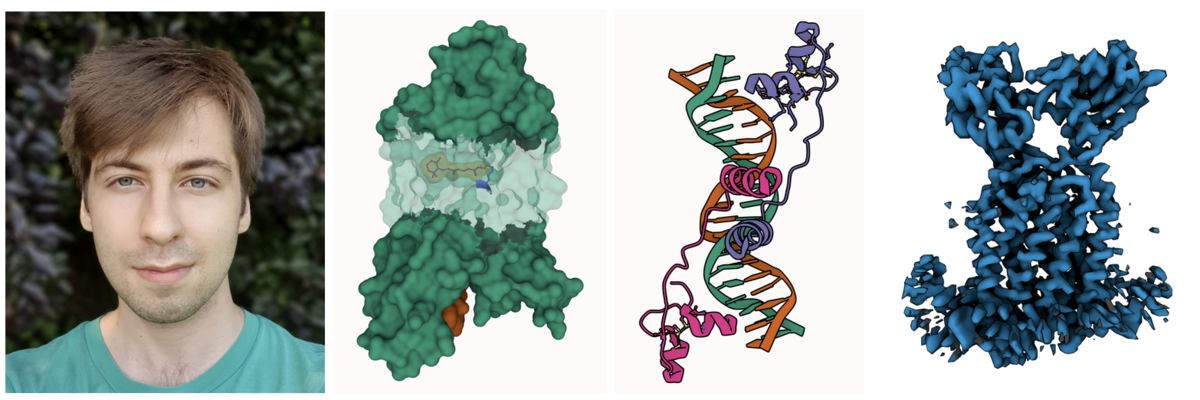 Photo of Áron; Partial transparency revealing retinal and surrounding channels in 3PQR; Stylized depiction of PDB ID 1D66 with crisp outlines; and Cryo-EM density of EMD-12604 with shadowing of occluded crevices.
Photo of Áron; Partial transparency revealing retinal and surrounding channels in 3PQR; Stylized depiction of PDB ID 1D66 with crisp outlines; and Cryo-EM density of EMD-12604 with shadowing of occluded crevices.Áron Samuel Kovács just finished his Master thesis in computer graphics at Masaryk University in the group of Barbora Kozlíková. He greatly improved the 3D rendering capabilities of Mol*, including artifacts-free transparency, improved darkening of crevices for better depth perception and much cleaner outlines.
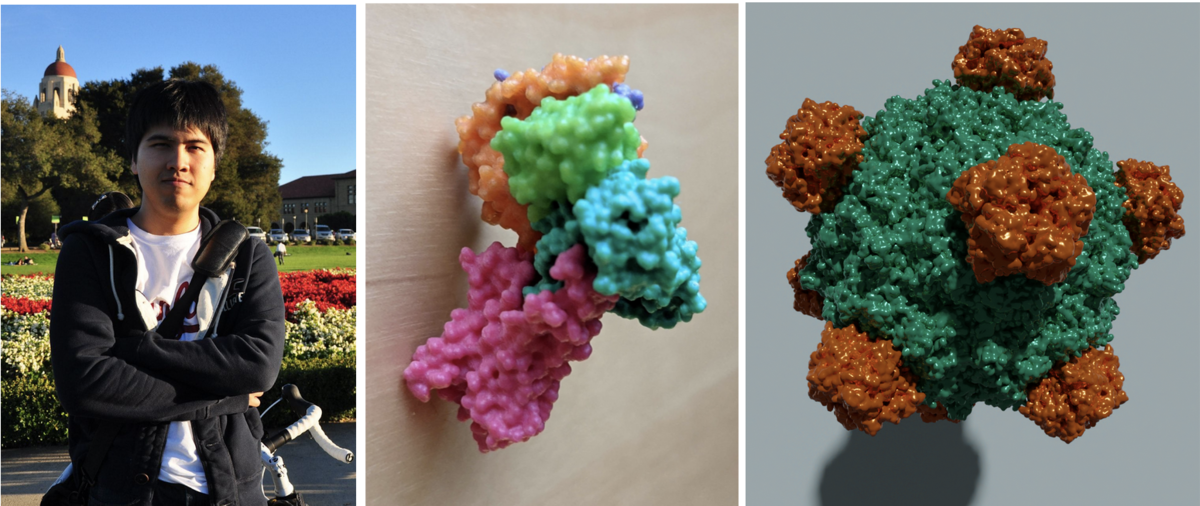 Photo of Sukolsak. 3D printed model of PDB ID 3SN6. High quality image of PDB ID 1RB8 rendered with the free and open source tool Blender.
Photo of Sukolsak. 3D printed model of PDB ID 3SN6. High quality image of PDB ID 1RB8 rendered with the free and open source tool Blender.Sukolsak Sakshuwong just finished his PhD in Management Science and Engineering at Stanford University in the group of Ashish Goel. He added geometry exporters to Mol* which allows users to extract 3D molecular scenes created in Mol* for use in 3D printing and other 3D graphic design. These scenes can be exported as glTF, an industry standard file format, as well as STL and Wavefront (.obj) formats.
The Mol* toolkit is available open access on GitHub, allowing community contributions.
06/24/2021
EMDB becomes a partner in wwPDB
The Electron Microscopy Data Bank (EMDB), the public repository for electron cryo-microscopy maps and tomograms of macromolecular complexes and subcellular structures, is now an official partner in the Worldwide Protein Data Bank (wwPDB) collaboration under a formal agreement.
The wwPDB partners are organizations that act as deposition, data processing and distribution centers for the three core wwPDB archives – Biological Magnetic Resonance Data Bank (BMRB), EMDB, and the Protein Data Bank (PDB).
The founding members--Research Collaboratory for Structural Bioinformatics PDB (RCSB PDB, USA), PDBe (Europe), and PDBj (Japan)--established the wwPDB in 2003. BMRB (USA) joined in 2006.
This move formalizes a long-standing relationship between the EMDB and wwPDB. EMDB was established in 2002 at EMBL’s European Bioinformatics Institute (EMBL-EBI). Since then, wwPDB and EMDB have collaborated on a wide range of issues including data deposition, annotation, and validation.
The partnership marks an important milestone in the wwPDB’s mission to bring coherence to the public archiving, management and dissemination of structural biology data, and highlights its commitment to the FAIR Principles (Findability, Accessibility, Interoperability, Reusability), which are emblematic of responsible stewardship of public domain information.
Key benefits of the partnership for EMDB users include the streamlining and harmonisation of policies and practices with the other core wwPDB archives to facilitate deposition, as well as improvements to data validation, which will facilitate the reuse of EMDB data.
06/22/2021
PDB50 at ACS August 25
 The Fall 2021 ACS meeting will be hybrid.
The Fall 2021 ACS meeting will be hybrid.Celebrate PDB50 at the Fall 2021 ACS Meeting with a session on Understanding Enzyme Function in 3D: Celebrating 50 Years of the Protein Data Bank.
All times shown are are listed in Eastern Daylight Time (EDT) on Wednesday August 25, 2021.
2:00 Introductory Remarks
2:05 The winding road from G-quadruplexes to telomerase, Juli Feigon (UCLA)
2:45 Enhanced exploration of small-molecule ligands bound to proteins and nucleic acids, Stephen K. Burley (Rutgers University and UCSD)
3:10 Mechanistic insights into the cleavage and polyadenylation machinery, Lori Passmore (University of Cambridge)
3:35 Vive la difference! The synergies and differences between the PDB and the CSD, Jason Cole (CCDC)
4:00 Break
4:30 Beyond static snapshots of protein structure: The role of dynamics in function, George Phillips Jr. (Rice University)
5:10 Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the COVID-19 pandemic, Sagar Khare (Rutgers)
5:35 Cracking the phosphatase code: Holoenzyme formation, regulatory protein binding and susbtrate dephosphorylation by the phosphoprotein phosphatase family, Rebecca Page (Brown)
6:00 Small molecules targeting COVID-19 in an evolving landscape of publishing and peer review, James Fraser (UCSF)
6:30 Break
7:00 Watching metalloenzymes at work, Amie Boal (Penn State)
7:25 Time travel to the past and future – evolution of energy landscapes for enzymes catalysis, Dorothee Kern (Brandeis)
7:50 Structure, mechanism, and inhibition of class IIb histone deacetylases, David Christianson (Harvard)
8:15 Truth sometimes triumphs: The history of structural enzymology, Gregory Petsko (Brandeis)
8:55 Closing Remarks
Visit ACS for registration information.
This symposium was organized by Carmen Nitsche (CCDC), and Steven C. Almo (Albert Einstein College of Medicine), and Stephen K. Burley (RCSB PDB).
06/08/2021
Congratulations to Poster Prize Winners
 Poster Prize awardees
Poster Prize awardeesAt the inaugural PDB50 meeting, ~275 posters were presented (Abstracts Day 1 | Day 2); 209 of these presentations were considered for poster prize awards.
- Best in High School: Nicholas Mamisashvili, Shelter Island High School, Molecular Dynamics Simulation of 6PEY.pdb a Novel Mutation in the Enzyme Methylenetetrahydrofolate Reductase
- Best in Undergraduate: Ijeoma Okoye, Vassar College, X-ray and Antioxidant Determination of Butein and 2’,4’-dihydroxy-3,4-dimethoxychalcone to Examine their Antimalarial Activity by Binding to Falcipain-2
- Best in Graduate: Daniel Sultanov, New York University, Mining for functional ribosomal variants in Saccharomyces cerevisiae
- Best in Postdoctoral Scholars: Seda Kocaman, National Institute of Environmental Health Sciences, Different ATP binding states of the essential AAA (ATPases Associated with various Activities)-ATPase Rix7 facilitate substrate translocation in ribosome biogenesis
Many thanks to the poster prize judges:
- BMRB: Hamid Eghbalnia
- PDBe: Genevieve Evans, John Berrisford
- PDBj: Genji Kurisu
- UConn: Bing Hao, Irina Bezsonova, Melissa Caimano
- University of Naples: Luigi Di Costanzo
- RCSB: Brian Hudson, Brinda Vallat, Cathy Lawson, Chenghua Shao,
- David Goodsell, Dennis Piehl, Ezra Peisach, Helen Berman, Irina Persikova, Joan Segura, Justin Flatt, Rachel Kramer Green, Stephen Burley, Yuhe Liang, Zukang Feng
- RIT: Paul Craig
wwPDB is celebrating the 50th Anniversary of the PDB throughout 2021 with symposia, materials, and more.
06/01/2021
Consistent Format for Validation and Coordinate Data
wwPDB validation reports are now provided in PDBx/mmCIF format for all new depositions in OneDep. This change makes validation data more interoperable with the PDB archival format. Data are more logically and better organized in the PDBx/mmCIF reports, and therefore more “database-friendly” than the report in XML format. PDBx/mmCIF-format validation reports for newly released and modified entries will be distributed through the PDB and EMDB Core Archives.
The new PDBx/mmCIF reports are easier to interpret. They contain a high-level summary and offer easier access to residue-level information. Data are provided at multiple levels: entity, chain-specific, and even at the individual residues. For example, it is more straightforward to obtain the total number of clashes. The corresponding validation dictionary is available at mmcif.wwpdb.org/dictionaries/mmcif_pdbx_vrpt.dic/Index. Examples of PDBx/mmCIF validation reports for X-ray, 3DEM, and NMR are publicly available at GitHub.
PDBx/mmCIF validation reports will be provided for the full PDB and EMDB archives once archival validation recalculation is performed.
wwPDB strongly recommends all PDB users and software developers adopt this format for future applications.
05/30/2021
Modifications to support for SHEET and ligand SITE records in June 2021
In 2014, PDBx/mmCIF became the PDB’s archive format and the the legacy PDB file format was frozen. In addition to PDBx/mmCIF files for all entries, wwPDB produces PDB format-formatted files for entries that can be represented in this legacy file format (e.g., entries with over 99,999 atoms or with multi-character chain IDs are only available in PDBx/mmCIF).
As the size and complexity of PDB structures increases, additional limitations of the legacy PDB format are becoming apparent and need to be addressed (as announced previously).
Defining complex sheet records
Restrictions in the SHEET record fields in legacy the PDB file format do not allow for the generation of complex beta sheet topology. Complex beta sheet topologies include instances where beta strands are part of multiple beta sheets and other cases where the definition of the strands within a beta sheet cannot be presented in a linear description. For example, in PDB entry 5wln a large beta barrel structure is created from multiple copies of a single protein; within the beta sheet forming the barrel are instances of a single beta strand making contacts on one side with multiple other strands, even from different chains.
This limitation, however, is not an issue in the PDBx/mmCIF formatted file, where these complex beta sheet topology can be captured in _struct_sheet, _struct_sheet_order, _struct_sheet_range, and _struct_sheet_hbond.
Starting June 8th 2021, legacy PDB format files will no longer be generated for PDB entries where the SHEET topology cannot be generated. For these structures, wwPDB will continue to provide secondary structure information with helix and sheet information in the PDBx/mmCIF formatted file.
Deprecation of _struct_site (SITE) records
wwPDB regularly reviews the software used during OneDep biocuration. The _struct_site and _struct_site_gen categories in PDBx/mmCIF (SITE records in the legacy PDB file format) are generated by in-house software and based purely upon distance calculations, and therefore may not reflect biological functional sites.
Starting in June 2021, the in-house legacy software which produces _struct_site and _struct_site_gen records will be retired and wwPDB will no longer generate these categories for newly-deposited PDB entries. Existing entries will be unaffected.
05/25/2021
How the Protein Data Bank Changed Biology
Journal of Biological Chemistry (JBC) has published a collection of reviews in celebration of PDB50.
This issue, edited by Lila Gierasch (JBC) and Helen Berman (wwPDB Foundation, RCSB PDB), contains 17 reviews highlighting the impact of the PDB archive across biological chemistry.
JBC was one of the first journals to require PDB deposition of structural data reported in accepted articles. In addition, more structures in the PDB have been published in JBC than in any other journal.
04/27/2021
Fifty years of collaborative science
If the past 15 months have taught us anything about science, it’s that it is vital for researchers to work together to make progress on major challenges. Scientists from around the world will come together virtually to celebrate the 50th anniversary of a key piece of the infrastructure for sharing scientific knowledge: the Protein Data Bank (PDB). The event will be hosted by the American Society for Biochemistry and Molecular Biology on May 4–5, 2021.
Additional events and resources will be announced throughout the year at wwpdb.org/pdb50.
The PDB is the global archive for biological structures. From its inception, the PDB has embraced a culture of open access, leading to its widespread use by the research community and public alike. Millions of users access the PDB data exploring fundamental biology, energy and biomedicine.
Structural biology archived in the PDB opens windows into biology. Through their structures, scientists not only can understand how biological molecules work but can design many of our modern medicines.
Structural biology has been seminal in understanding how SARS-CoV-2, the virus that causes COVID-19, and is the foundation of our understanding of protein folding. In fact, more structural biologists have been awarded Nobel Prizes than those in any other field.
In 1971, Helen Berman, a co-founder of the PDB and now a professor emerita at Rutgers University, and colleagues realized that the research community would benefit from sharing structural biology data. The PDB archive that they started has grown into a global database managed by the Worldwide Protein Data Bank consortium (wwPDB) of partner sites in Asia, Europe and America.
“The PDB plays a seminal role in structure-based drug design, a mainstay of many of our current therapeutics… (and) has given rise to the entire field of structural bioinformatics,” Berman said.
Most scientific journals require deposition of structural biology data in the PDB prior to publication. The PDB data are readily accessible to scientists, educators and nonscientists alike.
Leading structural biologists at the meeting from Caltech, Stanford University, Tsinghua University, Harvard Medical School and many other institutions will celebrate the history of the PDB archive. They will also present their current research on topics ranging from SARS-CoV-2 replication, cancer therapies based on antibodies conjugated to small molecules, and immunity and antiviral drugs.
Thousands of scientists have contributed and access the PDB archive regularly. The Journal of Biological Chemistry recently released a special issue on this theme, scientific advances enabled by the PDB.
The importance of sharing structural biology data for systems biology, protein design and drug discovery will continue to open our world into the intricacies biology.
Learn more about the upcoming May meeting and review the agenda at ASBMB. Registration ends May 1.
04/20/2021
Future Planning: Entries with extended PDB and CCD ID codes will be distributed in PDBx/mmCIF format only
wwPDB, in collaboration with the PDBx/mmCIF Working Group, has set plans to extend the length of ID codes for PDB and Chemical Component Dictionary (CCD) ID entries in the future. Entries containing these extended IDs will not be supported by the legacy PDB file format.
CCD entries are currently identified by unique three-character alphanumeric codes. At current growth rates, we anticipate running out of available new codes in the next three to four years. At this point, the wwPDB will issue four-character alphanumeric codes for CCD IDs in the OneDep system. Due to constraints of the legacy PDB file format, entries containing these new, four character ID codes will only be distributed in PDBx/mmCIF format. The wwPDB will begin implementation of extended CCD ID codes in 2022.
In addition, wwPDB also plans to extend PDB ID length to eight characters prefixed by ‘PDB’, e.g., pdb_00001abc. Each PDB ID has a corresponding Digital Object Identifier (DOI), often required for manuscript submission to journals and described in publications by the structure authors. Both extended PDB IDs and corresponding PDB DOIs, along with existing four character PDB IDs, will be included in the PDBx/mmCIF formatted files for all new entries by Fall 2021.
For example, PDB entry 1ABC will also have the extended PDB ID (pdb_00001abc) and the corresponding PDB DOI (10.2210/pdb1abc/pdb) listed in the _database_2 PDBx/mmCIF category.
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB 1abc pdb_00001abc 10.2210/pdb1abc/pdb
WWPDB D_1xxxxxxxxx ? ?
Once four-character PDB IDs are all consumed, newly-deposited PDB entries will only be issued extended PDB ID codes, and entries will only be distributed in PDBx/mmCIF format.
wwPDB is asking PDB users and related software developers to review code and begin to remove such limitations for the future.
04/18/2021
Register for PDB50 by May 1
Throughout 2021, the wwPDB will be celebrating the 50th anniversary of the PDB archive (wwpdb.org/pdb50).
The inaugural symposium will be held virtually on May 4-5, 2021.
The online sessions will take place between 11 a.m. – 4:30 p.m. ET each day. The event will be recorded and made available to registered participants after the meeting.
Students and postdoctoral fellows are especially encouraged to attend and will be eligible for poster awards.
Register by May 1 at https://www.asbmb.org/meetings-events/pdb50.
Speakers
04/09/2021
New pdb-l Bulletin Board
pdb-l@lists.wwpdb.org is an open discussion forum for questions and discussions with the PDB user community about protein structure, analysis, and related topics. Messages sent to pdb-l@lists.wwpdb.org will be sent to all subscribers. This bulletin board, which supports HTML formatting and images, replaces the previous forum at pdb-l@sdsc.edu. Messages, including messages migrated from the previous bulletin board are archived.
Existing subscribers do not need to resubscribe. New users should visit the pdb-l@lists.wwpdb.org info page to subscribe.
Questions about the migration should be sent to info@wwpdb.org.
Questions about PDB structures should be sent to deposit-help@mail.wwpdb.org.
04/06/2021
Biocurator Milestone: >10,000 Depositions Processed
Congratulations to wwPDB’s Ms. Yumiko Kengaku on processing 10,000 depositions. Yumiko began her career as a biocurator in 2000, as a member the newly-formed PDBj team at her alma mater Osaka University. She is the 1st wwPDB biocurator to process more than 10,000 structures. Many Asian structural biologists know and trust Yumiko. For more than two decades, she has worked closely with depositors to expertly guiding them through the structure deposition process, ensuring timely release of high-quality data. A gift celebrating her long service to PDBj, the wwPDB, and the global scientific community was presented to Yumiko in April 2021. We look forward to celebrating the accomplishments of the next biocurator to reach the 10,000 deposition milestone.
 PDBj Head Genji Kurisu and Ms. Yumiko Kengaku
PDBj Head Genji Kurisu and Ms. Yumiko Kengaku
03/31/2021
Improved support for extended PDBx/mmCIF structure factor files
Extensions to the PDBx/mmCIF dictionary for reflection data with anisotropic diffraction limits, for unmerged reflection data, and for quality metrics of anomalous diffraction data are now supported in OneDep.
In October 2020, a subgroup of the wwPDB PDBx/mmCIF Working Group was convened to develop a richer description of experimental data and associated data quality metrics. Members of this Data Collection and Processing Subgroup are all actively engaged in development and support of diffraction data processing software. The Subgroup met virtually for several months discussing, reviewing, and finalizing a new set dictionary content extension that were incorporated into the PDBx/mmCIF dictionary on February 16, 2021. A reference implementation of the new content extensions has been developed by Global Phasing Ltd.
These extensions facilitate the deposition and archiving of a broader range of diffraction data, as well as new quality metrics pertaining to these data. These extensions cover three main areas:
- scaled and merged reflection data that have been processed to take account of diffraction anisotropy, by providing descriptors for that anisotropy, in terms of (1) a parameter-free definition of a cut-off surface by means of a per-reflection “signal” and a threshold value for that signal, and (2) the ellipsoid providing the best fit to the resulting cut-off surface;
- scaled and unmerged reflection data, by providing extra item definitions aimed at ensuring that such data can be meaningfully re-analysed, and their quality assessed independently from the associated model, after retrieval from the archive;
- anomalous diffraction data, by adding descriptors for numerous relevant, but previously missing, statistics.
The new mmCIF data extensions describing anisotropic diffraction now enable archiving of the results of Global Phasing’s STARANISO program. Developers of other software can make use of them or extend the present definitions to suit their applications. Example files created by autoPROC, BUSTER (version 20210224) and Gemmi that are compliant with the new dictionary extensions are provided in a GitHub repository.
These example files, and similarly compliant files produced by other data processing and/or refinement programs, are suitable for direct uploading to the wwPDB OneDep system. Automatic recognition of that compliance, implemented by means of explicit dictionary versioning using the new pdbx_audit_conform record, will avoid unnecessary pre-processing at the time of deposition. This improved OneDep support will ensure a lossless round trip between data processing/refinement in the lab and deposition at the PDB.
wwPDB strongly encourages structural biologists to always use the latest versions of structure determination software packages to produce data files for PDB deposition. wwPDB also encourages crystallographers wishing to deposit new structures together with their associated diffraction data to use the software which guarantees consistency between data and final model. This consistency is difficult to achieve when separate diffraction data files and model coordinate files are pieced together a posteriori by ad hoc means.
wwPDB also encourages depositors to make their raw diffraction images available from one of the public repositories to allow direct access to the original diffraction image data.
03/29/2021
OneDep highlights curated assemblies for review in Mol*
To improve the clarity of assembly definitions in curation, wwPDB now makes curated PDB assemblies available for depositors to view in OneDep using the Mol* viewer.
One of the important processes in curation of PDB entries is the definition of assemblies for each structure. This helps users of PDB data to understand the structure in the context of its complex formation in the specific experimental conditions.
To ensure that assemblies are curated correctly, they are reviewed by annotators at the time of curation before being reported back to the depositors after the curation process.
The deposition system in OneDep has now been enhanced so that after curation, the annotated assembly is displayed in the Mol* 3D viewer for depositors to review. This viewer is available in a new Review section in the deposition interface, which is present after curation of the entry. The Mol* viewer can display PDB structure data within the browser with minimal memory requirements, therefore making it quick and easy to visually display assembly information.
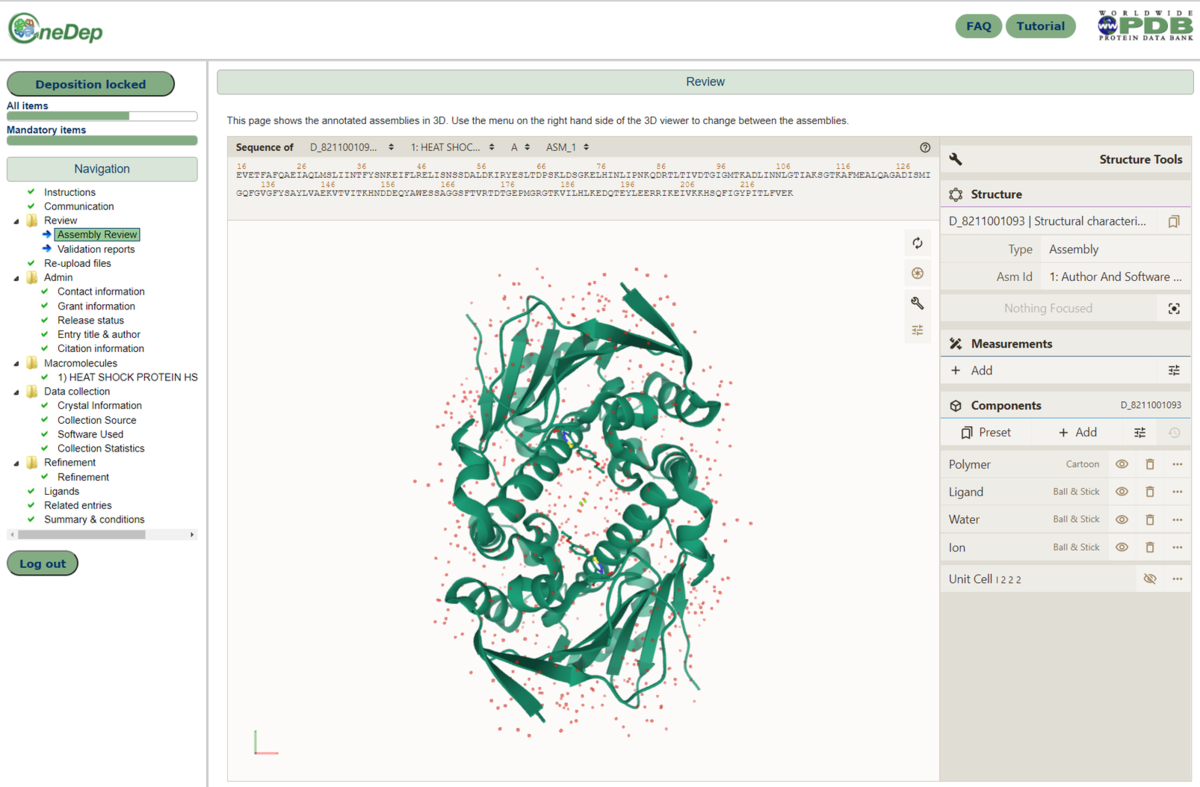 The assembly review page, as displayed for depositors after curation of the entry. The curated assembly is displayed in the Mol* 3D viewer, within the browser.
The assembly review page, as displayed for depositors after curation of the entry. The curated assembly is displayed in the Mol* 3D viewer, within the browser.These changes will help improve the validation and reporting of curated assemblies during the deposition process.
03/16/2021
Modifications to support for SHEET and ligand SITE records in June 2021
In 2014, PDBx/mmCIF became the PDB’s archive format and the the legacy PDB file format was frozen. In addition to PDBx/mmCIF files for all entries, wwPDB produces PDB format-formatted files for entries that can be represented in this legacy file format (e.g., entries with over 99,999 atoms or with multi-character chain IDs are only available in PDBx/mmCIF)
As the size and complexity of PDB structures increases, additional limitations of the legacy PDB format are becoming apparent and need to be addressed.
Defining complex sheet records
Restrictions in the SHEET record fields in legacy the PDB file format do not allow for the generation of complex beta sheet topology. Complex beta sheet topologies include instances where beta strands are part of multiple beta sheets and other cases where the definition of the strands within a beta sheet cannot be presented in a linear description. For example, in PDB entry 5wln a large beta barrel structure is created from multiple copies of a single protein; within the beta sheet forming the barrel are instances of a single beta strand making contacts on one side with multiple other strands, even from different chains.
This limitation, however, is not an issue in the PDBx/mmCIF formatted file, where these complex beta sheet topology can be captured in _struct_sheet, _struct_sheet_order, _struct_sheet_range, and _struct_sheet_hbond.
Starting June 8th 2021, legacy PDB format files will no longer be generated for PDB entries where the SHEET topology cannot be generated. For these structures, wwPDB will continue to provide secondary structure information with helix and sheet information in the PDBx/mmCIF formatted file.
Deprecation of _struct_site (SITE) records
wwPDB regularly reviews the software used during OneDep biocuration. The _struct_site and _struct_site_gen categories in PDBx/mmCIF (SITE records in the legacy PDB file format) are generated by in-house software and based purely upon distance calculations, and therefore may not reflect biological functional sites.
Starting in June 2021, the in-house legacy software which produces _struct_site and _struct_site_gen records will be retired and wwPDB will no longer generate these categories for newly-deposited PDB entries. Existing entries will be unaffected.
03/15/2021
Enhanced Validation of Small-Molecule Ligands and Carbohydrates
A new article in Structure describes new features, including branched representations and 2D SNFG images for carbohydrates, identification of ligands of interest, 3D views of electron density fit, and 2D images of small molecule geometry.
These enhancements and processes for validation of 3D small-molecular structures reflect recommendations from the wwPDB/CCDC/D3R Ligand Validation Workshop and the adoption of software through community collaborations.
This manuscript also highlights enhancements made since the initial implementation of Validation Reports as described in Validation of the Structures in the Protein Data Bank (2017) Structure 25: 1916-1927 doi: 10.1016/j.str.2017.10.009.
Enhanced Validation of Small-Molecule Ligands and Carbohydrates in the Protein Data Bank
Zukang Feng, John D. Westbrook, Raul Sala, Oliver S. Smart, Gérard Bricogne, Masaaki Matsubara, Issaku Yamada, Shinichiro Tsuchiya, Kiyoko F. Aoki-Kinoshita, Jeffrey C. Hoch, Genji Kurisu, Sameer Velankar, Stephen K. Burley, and Jasmine Y. Young
(2021) Structure doi: 10.1016/j.str.2021.02.004
03/08/2021
Submit Abstracts for PDB50
Throughout 2021, the wwPDB will be celebrating the 50th anniversary of the PDB archive (wwpdb.org/pdb50).
The inaugural symposium will be held virtually on May 4-5, 2021.
The online sessions will take place between 11 a.m. – 4:30 p.m. ET each day. The event will be recorded and made available to registered participants after the meeting.
Students and postdoctoral fellows are especially encouraged to attend and will be eligible for poster awards.
Abstract submission and reduced registration rates end March 22. Register at https://www.asbmb.org/meetings-events/pdb50.
Speakers
03/02/2021
More than 1,000 SARS-CoV-2 Coronavirus Protein Structures Available
With this week's update, 1,018 SARS-CoV-2-related structures are now freely available from the Protein Data Bank.
The first SARS-CoV-2 structure, a high-resolution crystal structure of the coronavirus main protease (PDB 6lu7), was released early in the pandemic on February 5, 2020.
Since then, structural biologists have visualized most of the SARS-CoV-2 proteome, including the spike protein binding to its ACE2 receptor and neutralizing antibodies, and the main protease, the papain-like proteinase, and other promising drug discovery targets. All of the structures and related data are available for exploration from wwPDB partner websites: RCSB PDB, PDBe, PDBj, and BMRB.
Rapid public release of SARS-CoV-2 structure data has greatly increased our understanding of Covid-19, allowed direct visualization of emerging variants of the virus, and facilitated structure-guided drug discovery and reuse to combat infection. Open access to PDB structures has already enabled design of effective vaccines against SARS-CoV-2.
The response of the research community to the pandemic has highlighted the importance of open access to scientific data in real time. The wwPDB strives to ensure that 3D biological structure data remain freely accessible for all, while maintaining as comprehensive and accurate an archive as possible.
The impact of these 1018 structures and many more coronavirus protein structures to come stands as a testament to the importance of open access to structural biology research data.
01/14/2021
PDB50: Registration Open for Virtual Event
Throughout 2021, the wwPDB will be celebrating the 50th anniversary of the PDB archive.
The inaugural symposium will be held May 4-5, 2021 in an event hosted by the American Society for Biochemistry and Molecular Biology and organized by the wwPDB Foundation.
This celebration of the 50th anniversary of the founding of the Protein Data Bank as the first open access digital data resource in biology will include presentations from speakers from around the world who have made tremendous advances in structural biology and bioinformatics.
Attendees are encouraged to participate in the virtual poster session and exhibition hall. Students and postdoctoral fellows will be eligible for poster awards.
Register and submit abstracts by March 15th, 2021 for reduced rates.
Speakers will include:
- Edward Arnold - Rutgers, The State University of New Jersey
- Helen M. Berman - Rutgers, The State University of New Jersey and University of Southern California
- Thomas L. Blundell - University of Cambridge
- Alexandre M. J. J. Bonvin - Utrecht University
- Stephen K. Burley - Rutgers, The State University of New Jersey and University of California San Diego
- Wah Chiu - Stanford University
- Johann Deisenhofer - University of Texas Southwestern Medical Center
- Juli Feigon - University of California Los Angeles
- Angela M. Gronenborn - University of Pittsburgh
- Jennifer L. Martin - University of Wollongong
- Stephen L. Mayo - California Institute of Technology
- Zihe Rao - ShanghaiTech University and Tsinghua University
- Hao Wu - Boston Children's Hospital and Harvard Medical School
The online sessions will take place between 11 a.m. – 4:30 p.m. EST each day. The event will be recorded and made available to registered participants after the meeting.
Sponsorship opportunities are available; please contact the wwPDB Foundation for more information.
01/12/2021
wwPDB EM Validation Reports Now Publicly Available
The wwPDB archive has now been updated to include validation reports for every released set of EM model coordinates in the PDB and every released EMDB map entry. Validation reports provide quantitative and visual assessments of structure quality and enable archive-wide comparisons (https://www.wwpdb.org/validation/validation-reports).
wwPDB EM validation reports were first made available to OneDep depositors in 2019 (http://www.wwpdb.org/news/news?year=2019#5db841ceea7d0653b99c8839). The current reports are based on recommendations obtained from EM Validation Task Force (VTF) meetings in 2010 (Structure 20: 205-214, wwpdb.org/task/em) and 2020 (white paper in preparation), as well as EM Validation Challenge events (https://www.ncbi.nlm.nih.gov/pubmed/32002441, https://www.biorxiv.org/content/10.1101/2020.06.12.147033v1). Examples of recent improvements include images for deposited masks, improved map-model overlay images, visualization of a (approximate) raw map from two half-maps, and rotationally averaged power spectrum plots. The underlying methodology is continually improved, based on community requirements, requests and feedback.
The PDB Core Archive holds validation reports that assess each PDB model along with its associated experimental map/tomogram from EMDB. EM map+model reports can be downloaded at the following wwPDB mirrors:
The EMDB Core Archive holds validation reports that assess each EMDB map/tomogram entry. EM map-only reports can be downloaded at the following URLs:
Additional information about validation reports is available for EM map+model, EM map only, and EM tomograms.
If you have any questions or queries about wwPDB validation, please contact us at validation@mail.wwpdb.org.
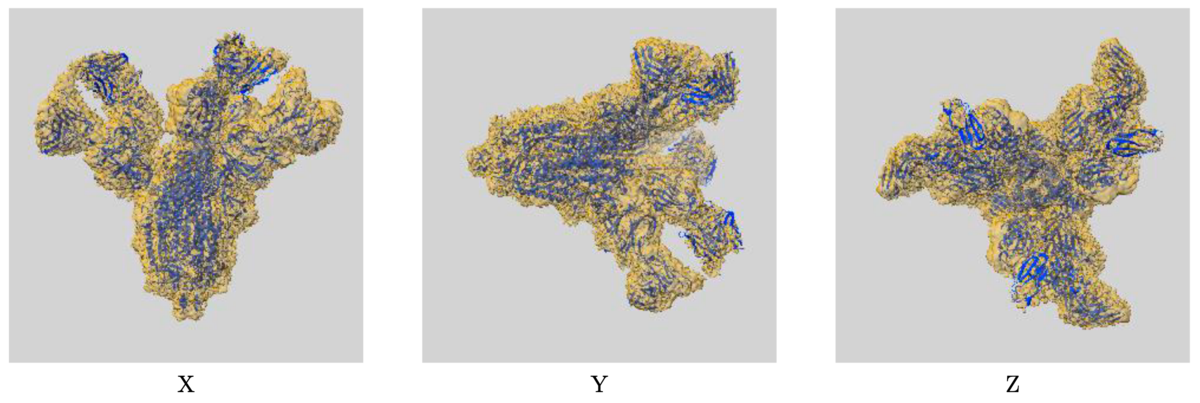 Example of map-model overlay image: EMD-30388/7CWU, SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail
Example of map-model overlay image: EMD-30388/7CWU, SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail
01/06/2021
Time-stamped Copies of wwPDB Archives
A snapshot of the PDB Core archive (ftp://ftp.wwpdb.org) as of January 5th, 2021 has been added to ftp://snapshots.wwpdb.org and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20210105 includes the structure and experimental data for the 173,005 PDB entries available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot of PDB Core archive is 822 GB.
A snapshot of the EMDB Core archive (ftp://ftp.ebi.ac.uk/pub/databases/emdb/) as of January 4, 2021 can be found in ftp://ftp.ebi.ac.uk/pub/databases/emdb_vault/20210101/ and ftp://snapshots.pdbj.org/20210101/. The snapshot of EMDB Core archive contains map files and their metadata within XML files for both released and obsoleted entries (13731 and 142, respectively) and is 2.9 TB in size.